Many different types of cells in the intact organism elaborate cell membrane protrusions for a variety of functions. Neuronal cells, for example, produce mushroom-shaped spines that function as sites of synapse formation on the surfaces of the dendrites. Pancreatic beta cells and T cells produce microvilli that function as sites of ligand sensing. These tubular protrusions contain a dense core of actin filaments that function to generate force and maintain the membrane deformation. In addition, these tubular membrane compartments contain specific protein complexes involved in signaling cascades that serve to initiate downstream processes such as long-term potentiation in neurons and glucose-stimulated insulin secretion in pancreatic beta cells.
Interestingly, in most cell types, these membrane protrusions have been shown to be motile, i.e., they have the ability to translocate in the plane of the membrane. In the case of microvilli on pancreatic beta cells, the movement is directed and occurs over distances that are several times the diameter of individual microvilli. These observations raise important questions: What is the functional significance of this motility, what is the mechanism of movement, and how is structural integrity of microvilli maintained?
These questions touch on a variety of important problems in cell biology including cytoskeletal– membrane dynamics and ligand sensing at the cellular level. The microvillus represents a structural unit that has the ability to translocate in the fluid lipid bilayer. Membrane fluidity at the junction between the microvillus and the plasma membrane becomes important for movement of the microvillus as an integral unit. These unanswered questions have profound implications on how we understand the sensory function of structures like microvilli and how we design studies to understand complex diseases like type 2 diabetes.
Read the rest in the February 2019 ACSB Newsletter (pdf).
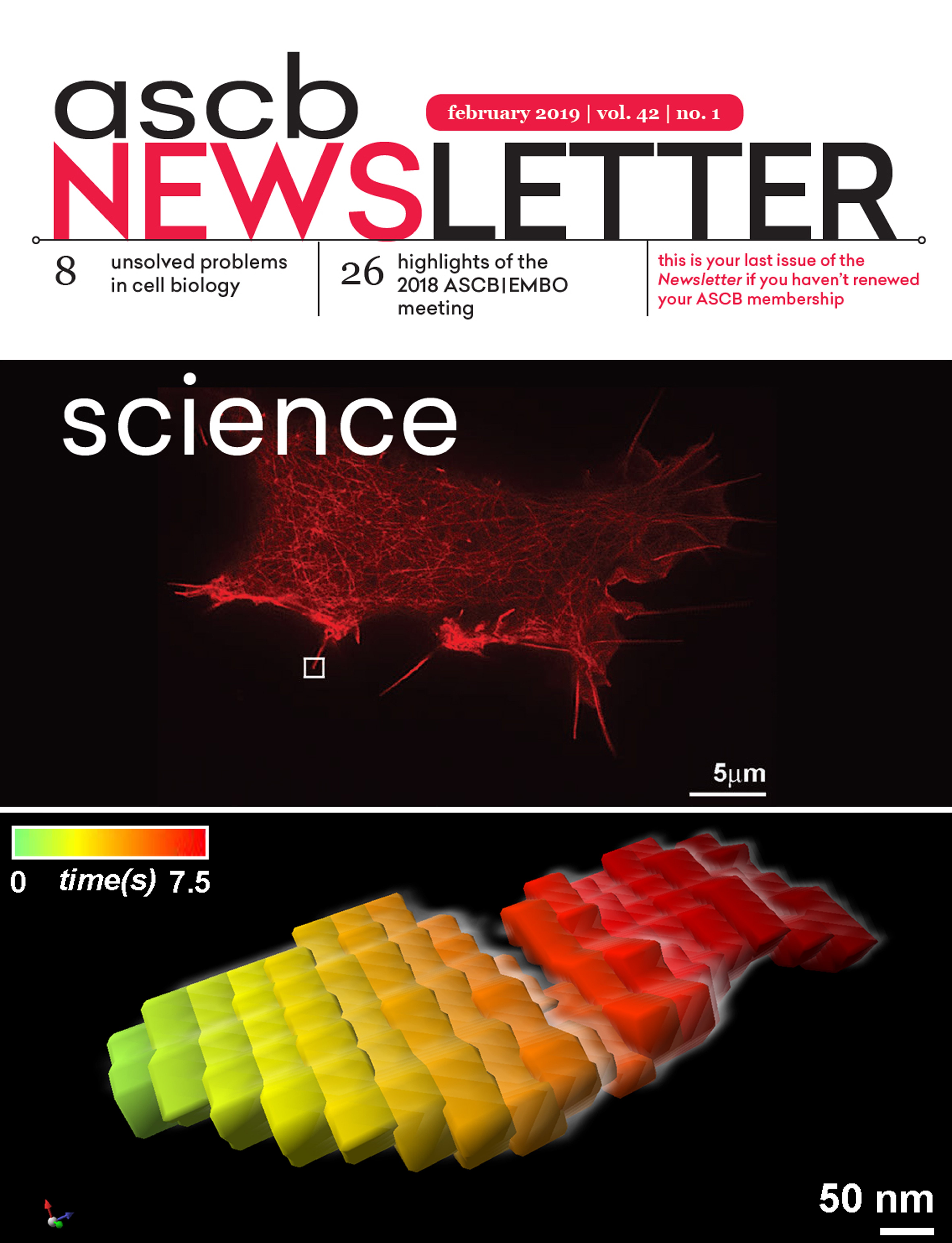
About the cover image: TIRF-SIM (Total internal reflection fluorescence structured illumination microscopy) live cell imaging of rat pancreatic beta cells (INS-1 832/13) transiently transfected with mRuby-LifeAct. The 3D kymograph in the bottom image shows the growing tip of a filopodium as it moved side-to-side over the 7.5-second time-lapse sequence. The Matlab function to generate 3D kymograph was written by Dr. Eric Wait, Advanced Imaging Center, HHMI Janelia Research Campus.
