
Eleanor Maine
- Biology
Research Interests
Genetic regulation of development, cell-signaling, germline development, RNA silencing.
Research Spotlight
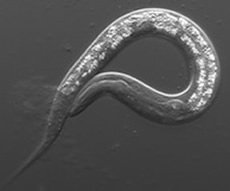
The Maine lab studies how cells and tissues form during animal development. Our work focuses on germline development in the model organism, Caenorhabditis elegans. We use a variety of genetic, molecular, genomics, and cell biological approaches to study germ cell proliferation and gamete formation. Currently, we focus on understanding RNA regulation that maintains germ cell identity and viability, ensures stem cell proliferation, and regulates chromatin structure during meiosis.
Degrees
- Postdoctoral Fellow, University of Wisconsin-Madison (1986-1990)
- Postdoctoral Fellow, Princeton University (1984-1986)
- Ph.D., Princeton University (1984)
Courses Taught
- BIO 305: Integrated Biology Laboratory
- BIO 700: Scientific Writing
Selected Publications
Caldas, I.V., L.H. Kelley, Y.H. Ahmed-Braimah, E.M. Maine (2023) smalldisco, a pipeline for siRNA discovery and 3’ tail identification. G3: Genes Genomes Genetics doi:10.1093/g3journal/jkad092
Brenner, J.L., E.M. Jyo^, A. Mohammad, P. Fox, V. Jones, E. Mardis, T. Schedl*, E.M. Maine* (2022) TRIM-NCL protein NHL-2 modulates cell fate choices in the C. elegans germ line. Developmental Biology 491: 43-55.
Vieux, K.-F., K. Prothro, L.H. Kelley, E.M. Maine, I. Veksler-Lublinsky, and K. McJunkin (2021) Screening by deep sequencing reveals mediators of miRNA tailing in C. elegans. bioRxiv preprint: doi.org/10.1101.2021.01.11.426275.
Li, Y., M. Snyder, and E.M. Maine (2021) Meiotic H3K9me2 distribution is influenced by the ALG-3 and ALG-4 pathway and by poly(U) polymerase activity. microPub Biology doi.org/10.18912/micropub.biology.000455.
Sullenberger, M.T., L.H. Kelley, and E.M. Maine (2020) PCR-based screening of small plasmid inserts. microPub Biology doi.org/10.17912/micropub.biology.000221.
Yang, B. , X. Xu, L. Russell, M.T. Sullenberger, J.L. Yanowitz, and E.M. Maine (2019) A DNA repair protein and histone methyltransferase interact to promote genome stability in C. elegans germ line. PLoS Genetics 15(2):e1007992. doi: 10.1371/journal.pgen.1007992.
Sullenberger, M.T., and E.M. Maine (2018) Simplified detection of a point mutation in C. elegans using tetra-primer ARMS-PCR. microPublication Biology doi.org/10.17912/MICROPUB.BIOLOGY.000078
Mutlu, B., H-M. Chen, J.J. Moresco, B.D. Orelo, B. Yang, J. Gaspar, S. Keppler-Ross, J.R. Yates III, D.H. Hall, E.M. Maine, and S.E. Mango (2018) Regulated nuclear accumulation of a histone methyltransferase times the onset of heterochromatin formation in C. elegans embryos. Science Advances 4: eaat6224. https://advances.sciencemag.org/content/4/8/eaat6224
Lissemore, J.L., E. Connors, Y. Liu, L. Qiao, B. Yang, M.L. Edgley, S. Flibotte, J. Taylor, V. Au, D.G. Moerman, and E.M. Maine (2018) The molecular chaperone HSP90 promotes Notch signaling in the germline of Caenorhabditis elegans. G3 8:1535-1544. https://doi.org/10.1534/g3.118.300551
Li, Y., and E.M. Maine (2018) The balance of poly(U) polymerase activity ensures germline identity, survival, and development in Caenorhabditis elegans. Development 145: dev165944. doi: 10.1242/dev.165944.
Safdar, K., A. Gu, X. Xu, V. Au, J. Taylor, S. Flibotte, D. G. Moerman, and E. M. Maine (2016) UBR-5, a conserved HECT-type E3 ubiquitin ligase, negatively regulates Notch-type signaling in C. elegans. G3 6:2125-2134.
Guo, Y., B. Yang, Y Li, X. Xu, and E. M. Maine (2015) Enrichment of H3K9me2 on unsynapsed chromatin in Caenorhabditis elegans does not target de novo sites. G3 5:1865-1878.
Van Wynsberghe, P. M., and E. M. Maine (2013) Epigenetic control of germline development. Advances in Experimental Medicine and Biology 757:373-403. Editor: T. Schedl. Publisher: Springer Verlag.
Fox, P.M., V.E. Vought, M. Hanazawa, M-H. Lee, E. M. Maine, and T. Schedl (2011) Cyclin E/CDK-2 regulates proliferative cell fate and cell cycle progression in the C. elegans germline. Development 138, 2223-2234.
Maine, E.M. (2010) Meiotic silencing in Caenorhabditis elegans. International Review of Cell and Molecular Biology 282, 91-134.
She, X., X. Xu, A. Fedotov, W.G. Kelly, and E.M. Maine (2009) Regulation of heterochromatin assembly during C. elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet 5(8): e1000624. doi:10.1371/journal.pgen.1000624
Maine, E.M. (2008) Studying gene function in Caenorhabditis elegans using RNA-mediated interference. Briefings in Functional Genomics and Proteomics 7, 184-194.
Liu, Y., and E.M. Maine (2007) The Bro1-domain protein, EGO-2, promotes notch signaling in Caenorhabditis elegans. Genetics 177, 2265-2277.
* This paper was featured in the Genetics Society of America Newsletter.
Maine, E. M., J. Hauth, T. Ratliff, V.E. Vought, X. She, and W. G. Kelly (2005) EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired DNA during C. elegans meiosis. Current Biology 15, 1972-1978.
* This paper was cited by the Faculty of 1000.
Smardon, A., J. M. Spoerke, S. C. Stacey, M. E. Klein, N. Mackin, and E. M. Maine (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germline development and RNA interference in C. elegans. Current Biology 10, 169-178.
* This paper was featured in Science Editor's Choice and Current Biology Dispatch reviews.
News
(May 11, 2022)
The prize memorializes William Wasserstrom, a noted professor of English at Syracuse University.
(Feb. 5, 2018)
Graduate student, Yini Li, identifies two proteins critical for development in the team’s study
(June 23, 2016)
Eleanor Maine Research Lab will study mechanisms, developmental importance of meiotic silencing
